Lapierre, É., Monthony, A. S. & Torkamaneh, D. Genomics-based taxonomy to explain hashish classification. Genome 66, 202–211 (2023).
Clarke, R. & Merlin, M. Hashish: Evolution and Ethnobotany. (Univ of California Press, 2016).
Hurgobin, B. et al. Fresh advances in Hashish sativa genomics analysis. New Phytol. 230, 73–89 (2021).
Cox, C. The Canadian Hashish Act legalizes and regulates leisure hashish use in 2018. Well being Coverage New York. 122, 205–209 (2018).
Welling, M. T. et al. A belated inexperienced revolution for Hashish: Digital genetic sources to fast-track cultivar construction. Entrance. Plant Sci. 7, 205761 (2016).
Martínez, V. et al. Cannabidiol and different non-psychoactive cannabinoids for prevention and remedy of gastrointestinal problems: helpful nutraceuticals?. Int. J. Mol. Sci. 21, 3067 (2020).
Torkamaneh, D. & Jones, A. M. P. Hashish, the multibillion greenback plant that no genebank sought after. Genome 65, 1–5 (2021).
Have an effect on at the Canadian financial system. (2023). To be had at: https://ised-isde.canada.ca/website online/competition-bureau-canada/en/how-we-foster-competition/education-and-outreach/planting-seeds-competition. (Accessed: fifth October 2023)
Hesami, M. et al. Fresh advances in hashish biotechnology. Ind. Plants Prod. 158, 113026 (2020).
Hanuš, L. O., Meyer, S. M., Muñoz, E., Taglialatela-Scafati, O. & Appendino, G. Phytocannabinoids: A unified crucial stock. Nat. Prod. Rep. 33, 1357–1392 (2016).
Sales space, J. Okay. & Bohlmann, J. Terpenes in Hashish sativa—From plant genome to people. Plant Sci. 284, 67–72 (2019).
Kovalchuk, I. et al. The genomics of hashish and its shut kinfolk. Annu. Rev. Plant Biol. 71, 713–739 (2020).
Grassa, C. J. et al. A brand new Hashish genome meeting friends increased cannabidiol (CBD) with hemp introgressed into marijuana. New Phytol. 230, 1665–1679 (2021).
Laverty, Okay. U. et al. A bodily and genetic map of Hashish sativa identifies intensive rearrangements on the THC/CBD acid synthase loci. Genome Res. 29, 146–156 (2019).
Grassa, C. J. et al. A whole Hashish chromosome meeting and adaptive admixture for increased cannabidiol (CBD) content material. bioRxiv. https://doi.org/10.1101/458083 (2018).
Gao, S. et al. A top of the range reference genome of untamed Hashish sativa. Hortic. Res. 7, 73. https://doi.org/10.1038/s41438-020-0295-3 (2020).
Maoz, T. Making hashish historical past in 2020. (2020). To be had at: https://nrgene.com/making-cannabis-history-in-2020/. (Accessed: sixth October 2023)
Morrell, P. L., Buckler, E. S. & Ross-Ibarra, J. Crop genomics: Advances and programs. Nat. Rev. Genet. 13, 85–96 (2012).
Schwabe, A. L. & McGlaughlin, M. E. Genetic gear weed out misconceptions of pressure reliability in Hashish sativa: Implications for a budding trade. J. Hashish Res. 1, 1–16 (2019).
Metzker, M. L. Sequencing applied sciences the following era. Nat. Rev. Genet. 11, 31–46 (2010).
Wang, J. & Zhang, Z. GAPIT model 3: Boosting energy and accuracy for genomic affiliation and prediction. Genom. Proteom. Bioinform. 19, 629–640 (2021).
Yin, L. et al. rMVP: A memory-efficient, visualization-enhanced, and parallel-accelerated software for genome-wide affiliation find out about. Genom. Proteom. Bioinform. 19, 619–628 (2021).
Torkamaneh, D. & Belzile, F. Genome-Extensive Affiliation Research. 2481, (Springer US, 2022).
Bakker, E., Holloway, A., Okay Waterman – US Patent App. 17/665, 500 & 2023, U. Autoflowering Markers. Google Patents (2021).
Welling, M. T. et al. An extreme-phenotype genome-wide affiliation find out about identifies candidate cannabinoid pathway genes in Hashish. Sci. Rep. 10, 1–14 (2020).
Music, Okay., Li, L. & Zhang, G. Protection advice for genotyping evaluation of extremely heterologous species the use of next-generation sequencing era. Sci. Rep. 6, 1–7 (2016).
Scariolo, F. et al. Genotyping evaluation by means of rad-seq reads comes in handy to evaluate the genetic id and relationships of breeding strains in lavender species aimed toward managing plant selection coverage. Genes (Basel). 12, 1656 (2021).
Sonah, H. et al. An advanced genotyping by means of sequencing (GBS) way providing greater versatility and potency of SNP discovery and genotyping. PLoS One 8, 1–9 (2013).
Torkamaneh, D., Laroche, J., Boyle, B., Hyten, D. L. & Belzile, F. A bumper crop of SNPs in soybean via high-density genotyping-by-sequencing (HD-GBS). Plant Biotechnol. J. 19, 860–862 (2021).
Poland, J. A. & Rife, T. W. Genotyping‐by means of‐sequencing for plant breeding and genetics. Plant Genome 5. https://doi.org/10.3835/plantgenome2012.05.0005 (2012).
Solar, J. et al. Genome-wide affiliation find out about of salt tolerance on the germination degree in hemp. Euphytica 219, 1–16 (2023).
Petit, J. et al. Elucidating the genetic structure of fiber high quality in hemp (Hashish sativa L.) the use of a genome-wide affiliation find out about. Entrance. Genet. 11, 566314. https://doi.org/10.3389/fgene.2020.566314 (2020).
Petit, J., Salentijn, E. M. J., Paulo, M. J., Denneboom, C. & Trindade, L. M. Genetic structure of flowering time and intercourse resolution in hemp (Hashish sativa L): A genome-wide affiliation find out about. Entrance. Plant Sci. 11, 569958 (2020).
Watts, S. et al. Hashish labelling is related to genetic variation in terpene synthase genes. Nat. Vegetation. 7, 1330–1334 (2021).
Collard, B. C. Y. & Mackill, D. J. Marker-assisted variety: An way for precision plant breeding within the twenty-first century. Philos. Trans. R. Soc. B Biol. Sci. 363, 557–572 (2008).
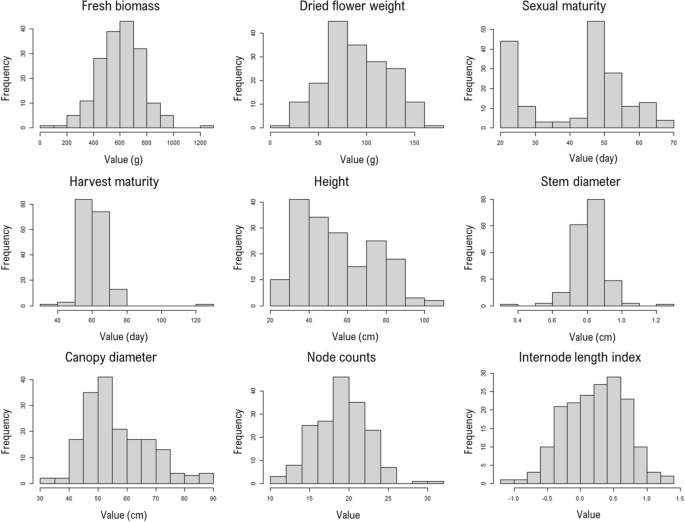
Lapierre, É., de Ronne, M., Boulanger, R. & Torkamaneh, D. Complete phenotypic characterization of numerous drug-type hashish types from the Canadian Prison Marketplace. Vegetation. 12, 3756 (2023).
Piluzza, G., Delogu, G., Cabras, A., Marceddu, S. & Bullitta, S. Differentiation between fiber and drug kinds of hemp (Hashish sativa L) from a number of wild and domesticated accessions. Genet. Resour. Crop Evol. 60, 2331–2342 (2013).
Jannink, J. L., Lorenz, A. J. & Iwata, H. Genomic variety in plant breeding: From idea to observe. Briefings Funct. Genom. Proteom. 9, 166–177 (2010).
Huang, M., Liu, X., Zhou, Y., Summers, R. M. & Zhang, Z. BLINK: A bundle for the following degree of genome-wide affiliation research with each people and markers within the thousands and thousands. Gigascience 8, 1–12 (2019).
R Core Group. R: The R Challenge for Statistical Computing. (2021). To be had at: https://www.r-project.org/. (Accessed: tenth June 2023)
Aboul-Maaty, N.A.-F. & Oraby, H.A.-S. Extraction of top of the range genomic DNA from other plant orders making use of a changed CTAB-based approach. Bull. Natl. Res. Cent. 43, 1–10 (2019).
Torkamaneh, D., Laroche, J. & Belzile, F. Speedy-gbs v2.0: An evaluation toolkit for genotyping-by-sequencing knowledge. Genome 63, 577–581 (2020).
Danecek, P. et al. The variant name layout and VCFtools. Bioinformatics 27, 2156–2158 (2011).
Browning, B. L. & Browning, S. R. Genotype imputation with thousands and thousands of reference samples. Am. J. Hum. Genet. 98, 116–126 (2016).
Danecek, P. et al. Twelve years of SAMtools and BCFtools. Gigascience 10, 1–4 (2021).
Bradbury, P. J. et al. TASSEL: Instrument for affiliation mapping of complicated characteristics in numerous samples. Bioinformatics 23, 2633–2635 (2007).
Quinlan, A. R. & Corridor, I. M. BEDTools: A versatile suite of utilities for evaluating genomic options. Bioinformatics 26, 841–842 (2010).
NCBI Hashish sativa Annotation Free up 100. (2019). To be had at: https://www.ncbi.nlm.nih.gov/genome/annotation_euk/Cannabis_sativa/100/. (Accessed: eleventh October 2023)
Nei, M. & Li, W. H. Mathematical type for finding out genetic variation when it comes to restriction endonucleases. Proc. Natl. Acad. Sci. 76, 5269–5273 (1979).
Purcell, S. et al. PLINK: A device set for whole-genome affiliation and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Remington, D. L. et al. Construction of linkage disequilibrium and phenotypic associations within the maize genome. Proc. Natl. Acad. Sci. USA. 98, 11479–11484 (2001).
Raj, A., Stephens, M. & Pritchard, J. Okay. FastSTRUCTURE: Variational inference of inhabitants construction in massive SNP knowledge units. Genetics 197, 573–589 (2014).
Jombart, T., Devillard, S. & Balloux, F. Discriminant evaluation of essential elements: A brand new approach for the evaluation of genetically structured populations. BMC Genet. 11, 1–15 (2010).
Emms, D. M. & Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 1–14 (2019).
Cheng, C. Y. et al. Araport11: A whole reannotation of the Arabidopsis thaliana reference genome. Plant J. 89, 789–804 (2017).
Torkamaneh, D. & Belzile, F. Scanning and filling: Extremely-dense SNP genotyping combining genotyping-by-sequencing, SNP array and whole-genome resequencing knowledge. PLoS One 10, e0131533 (2015).
Torkamaneh, D. et al. NanoGBS: A miniaturized process for GBS library preparation. Entrance. Genet. 11, 1–8 (2020).
Zhang, J. et al. Genome-wide affiliation find out about for flowering time, adulthood dates and plant peak in early maturing soybean (Glycine max) germplasm. BMC Genom. 16, 1–11 (2015).
Viana, J. P. G. et al. Have an effect on of more than one selective breeding systems on genetic range in soybean germplasm. Theor. Appl. Genet. 135, 1591–1602 (2022).
Liu, X., Geng, X., Zhang, H., Shen, H. & Yang, W. Affiliation and genetic identity of loci for 4 fruit characteristics in tomato the use of InDel markers. Entrance. Plant Sci. 8, 275267 (2017).
Ren, G. et al. Huge-scale whole-genome resequencing unravels the domestication historical past of Hashish sativa. Sci. Adv. 7, 2286–2302 (2021).
Torkamaneh, D. et al. Soybean (Glycine max) Haplotype Map (GmHapMap): A common useful resource for soybean translational and useful genomics. Plant Biotechnol. J. 19, 324–334 (2021).
Wang, W. et al. Genomic variation in 3010 numerous accessions of Asian cultivated rice. Nature. 557, 43–49 (2018).
Chia, J. M. et al. Maize HapMap2 identifies extant variation from a genome in flux. Nat. Genet. 44, 803–807 (2012).
Sawler, J. et al. The genetic construction of marijuana and hemp. PLoS One 10, e0133292 (2015).
Lynch, R. C. et al. Genomic and chemical range in Hashish. CRC. Crit. Rev. Plant Sci. 35, 349–363 (2016).
Soorni, A., Fatahi, R., Haak, D. C., Salami, S. A. & Bombarely, A. Overview of genetic range and inhabitants construction in Iranian Hashish Germplasm. Sci. Rep. 7, 1–10 (2017).
Zhang, J. et al. Genetic range and inhabitants construction of hashish in keeping with the genome-wide construction of straightforward collection repeat markers. Entrance. Genet. 11, 543438 (2020).
Clarke, R. C. & Merlin, M. D. Hashish domestication, breeding historical past, present-day genetic range, and long run potentialities. CRC. Crit. Rev. Plant Sci. 35, 293–327 (2016).
Lye, Z. N. & Purugganan, M. D. Replica quantity variation in domestication. Traits Plant Sci. 24, 352–365 (2019).
Springer, N. M. Epigenetics and crop development. Traits Genet. 29, 241–247 (2013).
Gill, R. A. et al. At the function of transposable parts within the law of gene expression and subgenomic interactions in crop genomes. CRC. Crit. Rev. Plant Sci. 40, 157–189 (2021).
Alonge, M. et al. Main affects of common structural variation on gene expression and crop development in tomato. (2020). https://doi.org/10.1016/j.mobile.2020.05.021
Liseron-Monfils, C. & Ware, D. Revealing gene law and associations via organic networks. Curr. Plant Biol. 3–4, 30–39 (2015).
Smýkal, P., Nelson, M. N., Berger, J. D. & Von Wettberg, E. J. B. The have an effect on of genetic adjustments all the way through crop domestication. Agronomy. 8, 119 (2018).
Schwabe, A. L., Hansen, C. J., Hyslop, R. M. & McGlaughlin, M. E. Comparative genetic construction of hashish sativa together with federally produced, wild amassed, and cultivated samples. Entrance. Plant Sci. 12, 675770 (2021).
de Ronne, M. et al. Mapping of partial resistance to Phytophthora sojae in soybean PIs the use of whole-genome sequencing finds a big QTL. Plant Genome 15, e20184 (2022).
Zhong, H. et al. Uncovering the genetic mechanisms regulating panicle structure in rice with GPWAS and GWAS. BMC Genom. 22, 1–13 (2021).
Cui, Z. et al. Denser markers and complex statistical approach recognized extra genetic loci related to husk characteristics in maize. Sci. Rep. 10, (2020).
Kaler, A. S., Gillman, J. D., Beissinger, T. & Purcell, L. C. Evaluating other statistical fashions and more than one trying out corrections for affiliation mapping in soybean and maize. Entrance. Plant Sci. 10, 1794. https://doi.org/10.3389/fpls.2019.01794 (2020).
Izquierdo, P., Kelly, J. D., Beebe, S. E. & Cichy, Okay. Mixture of meta-analysis of QTL and GWAS to discover the genetic structure of seed yield and seed yield elements in not unusual bean. Plant Genome 16, e20328 (2023).